Tutorial to Create a Discrete Event Simulation Model in R Using descem
This tutorial has been written by Javier Sanchez Alvarez and Valerie Aponte Ribero
Introduction
This document runs a discrete event simulation model in the context of early breast cancer to show how the functions can be used to generate a model in only a few steps.
When running a DES, it’s important to consider speed. Simulation based models can be computationally expensive, which means that using efficient coding can have a substantial impact on performance. Because the model is using parallel computing, in order to debug the model one can simply use browser() where needed and set the number of patients to be simulated equal to 1.
Main options
#devtools::install_github(
# "roche/Global-HTA-Evidence-Open",
# subdir = "Rpackages/descem"
# )
library(descem)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(flexsurv)
#> Loading required package: survival
library(ggplot2)
library(kableExtra)
#>
#> Attaching package: 'kableExtra'
#> The following object is masked from 'package:dplyr':
#>
#> group_rows
library(purrr)
library(tidyr)
options(scipen = 999)
options(tibble.print_max = 50)
General inputs with delayed execution
Initial inputs and flags that will be used in the model can be defined below. We can define inputs that are common to all patients (common_all_inputs) within a simulation, inputs that are unique to a patient independently of the treatment (e.g. natural death, defined in common_pt_inputs), and inputs that are unique to that patient and that treatment (unique_pt_inputs). Items can be included through the add_item function, and can be used in subsequent items. All these inputs are generated before the events and the reaction to events are executed. Furthermore, the program first executes common_all_inputs, then common_pt_inputs and then unique_pt_inputs. So one could use the items generated in common_all_inputs in unique_pt_inputs.
#Each patient is identified through "i"
#psa_bool is the indicator of whether we want to apply a PSA or not
#Put objects here that do not change on any patient or intervention loop
common_all_inputs <-add_item(
util.sick = 0.8,
util.sicker = 0.5,
cost.sick = 3000,
cost.sicker = 7000,
cost.int = 1000,
coef_noint = log(0.2),
HR_int = 0.8)
#Put objects here that do not change as we loop through treatments for a patient
common_pt_inputs <- add_item(death= max(0.0000001,rnorm(n=1, mean=12, sd=3)))
#Put objects here that change as we loop through treatments for each patient (e.g. events can affect fl.tx, but events do not affect nat.os.s)
unique_pt_inputs <- add_item(fl.sick = 1)
Events
Add Initial Events
Events are added below through the add_tte function. We use this function twice, one per intervention. We must define several arguments: one to indicate the intervention, one to define the names of the events used, one to define the names of other objects created that we would like to store (optional, maybe we generate an intermediate input which is not an event but that we want to save) and the actual input in which we generate the time to event. Events and other objects will be automatically initialized to Inf. We draw the times to event for the patients. This chunk is a bit more complex, so it’s worth spending a bit of time explaining it.
The init_event_list object is populated by using the add_tte function twice, one for the “int” strategy and other for the “noint” strategy. We first declare the start time to be 0.
We then proceed to generate the actual time to event. We use the draw_tte() function to generate the time to event. One should always be aware of how the competing risks interact with each other. While we have abstracted from these type of corrections here, it is recommended to have an understanding about how these affect the results and have a look at the competing risks/semi-competing risks literature.
init_event_list <-
add_tte(trt="noint", evts = c("sick","sicker","death") ,input={ #intervention
sick <- 0
sicker <- draw_tte(1,dist="exp", coef1=coef_noint)
}) %>% add_tte(trt="int", evts = c("sick","sicker","death") ,input={
sick <- 0
sicker <- draw_tte(1,dist="exp", coef1=coef_noint, hr = HR_int)
})
Add Reaction to Those Events
Once the initial times of the events have been defined, we also need to declare how events react and affect each other. To do so, we use the evt_react_list object and the add_reactevt function. This function just needs to state which event is affected, and the actual reaction (usually setting flags to 1 or 0, or creating new/adjusting events).
There are a series of objects that can be used in this context to help with the reactions. Apart from the global objects and flags defined above, we can also use curtime for the current event time, prevtime for the time of the previous event, cur_evtlist for the named vector of events that is yet to happen for that patient, trt for the current treatment in the loop, evt for the current event being processed, i expresses the patient iteration, and simulation the specific simulation (relevant when the number of simulations is greater than 1). Furthermore, one can also call any other input/item that has been created before or create new ones. For example, we could even modify a cost/utility item by changing it directly, e.g. through modify_item(list(cost.idfs.tx=500)).
| Item | What does it do |
|---|---|
curtime | Current event time (numeric) |
prevtime | Time of the previous event (numeric) |
cur_evtlist | Named vector of events that is yet to happen for that patient (named numeric vector) |
evt | Current event being processed (character) |
i | Patient being iterated (character) |
simulation | Simulation being iterated (numeric) |
The functions to add/modify events and inputs use lists. Whenever several inputs/events are added or modified, it’s recommended to group them within one function, as it reduces the computation cost. So rather than use two modify_item with a list of one element, it’s better to group them into a single modify_item with a list of two elements.
new_eventallows to generate events and add them to the vector of events. It accepts more than one event.
modify_event allows to modify events (e.g. delay death). When adding an event, the name of the events and the time of the events must be defined. When using modify_event, one must indicate which events are affected and what are the new times of the events. If the event specified does not exist or has already occurred, it will be ignored. Note that one could potentially omit part of the modeling set in init_event_list and actually define new events dynamically through the reactions (we do that below for the "ae" event). However, this can have an impact in computation time, so if possible it’s always better to use init_event_list.
modify_item allows to modify and add items. Elements defined within this function are not evaluated sequentially (i.e. defining modify_item(list(fl.new = 1, var1 = fl.new * 5))) will give an error if fl.new was not defined outside this function).
The list of relevant functions to be used within add_reactevt are:
| Function | What does it do | How to use it |
|---|---|---|
modify_item() | Adds & Modifies items/flags/variables for future events | modify_item(list("fl.idfs.ontx"=0,"fl.tx.beva"=0)) |
new_event() | Adds events to the vector of events for that patient | new_event(rep(list("ae"=curtime + 0.001),5)) |
modify_event() | Modifies existing events by changing their time | modify_event(list("os"=curtime +5, "ttot"=curtime+0.0001)) |
The model will run until curtime is set to Inf, so the event that terminates the model (in this case, os), should modify curtime and set it to Inf.
evt_react_list <-
add_reactevt(name_evt = "sick",
input = {}) %>%
add_reactevt(name_evt = "sicker",
input = {
modify_item(list(fl.sick = 0))
}) %>%
add_reactevt(name_evt = "death",
input = {
modify_item(list(curtime = Inf))
})
Costs and Utilities
Costs and utilities are introduced below. However, it’s worth noting that the model is able to run without costs or utilities.
Utilities
Utilities are defined using pipes with the add_util function. The first argument says which events are affected, the second argument which treatments are affected, and the third one describe the utilities. Instant utilities (e.g. AE disutilities) and cycle utilities can be defined in a similar fashion. Note that one can write expressions and objects whose execution will be delayed until the model runs.
util_ongoing <- add_util(evt = c("sick", "sicker","death"),
trt = c("int", "noint"),
util = util.sick * fl.sick + util.sicker * (1-fl.sick)
)
Costs
Costs are defined using pipes with the add_cost function, in a similar fashion to the utilities.
cost_ongoing <-
add_cost(
evt = c("sick", "sicker","death") ,
trt = "noint",
cost = cost.sick * fl.sick + cost.sicker * (1-fl.sick) ) %>%
add_cost(
evt = c("sick", "sicker","death") ,
trt = "int",
cost = cost.sick * fl.sick + cost.sicker * (1-fl.sick) + cost.int * fl.sick )
Model
Model Execution
The model can be run using the function RunSim below. We must define the number of patients to be simulated, the number of simulations, whether we want to run a PSA or not, the strategy list, the inputs, events and reactions defined above, the number of cores to be used (by default uses 1 core), the discount rate for costs and the discount rate for qalys. It is recommended not to use all the cores in the machine.
It is worth noting that the psa_bool argument does not run a PSA automatically, but is rather an additional input/flag of the model that we use as a reference to determine whether we want to use a deterministic or stochastic input. As such, it could also be defined in common_all_inputs as the first item to be defined, and the result would be the same. However, we recommend it to be defined in RunSim.
Note that the distribution chosen, the number of events and the interaction between events can have a substantial impact on the running time of the model.
#Logic is: per patient, per intervention, per event, react to that event.
results <- RunSim(
npats=1000, # number of patients to be simulated
n_sim=1, # number of simulations to run
psa_bool = FALSE, # use PSA or not. If n_sim > 1 and psa_bool = FALSE, then difference in outcomes is due to sampling (number of pats simulated)
trt_list = c("int", "noint"), # intervention list
common_all_inputs = common_all_inputs, # inputs common that do not change within a simulation
common_pt_inputs = common_pt_inputs, # inputs that change within a simulation but are not affected by the intervention
unique_pt_inputs = unique_pt_inputs, # inputs that change within a simulation between interventions
init_event_list = init_event_list, # initial event list
evt_react_list = evt_react_list, # reaction of events
util_ongoing_list = util_ongoing,
cost_ongoing_list = cost_ongoing,
ncores = 2, # number of cores to use, recommended not to use all
drc = 0.035, # discount rate for costs
drq = 0.035 # discount rate for qaly/lys
)
#> [1] "Simulation number: 1"
#> [1] "Time to run iteration 1: 0.47s"
#> [1] "Total time to run: 0.47s"
Post-processing of Model Outputs
Summary of Results
Once the model has been run, we can use the results and summarize them using the summary_results_det to print the results of the last simulation (if nsim=1, it’s the deterministic case), and summary_results_psa to show the PSA results (with the confidence intervals). We can also use the individual patient data generated by the simulation, which we collect here to plot in the psa_ipd object.
summary_results_det(results$final_output) #will print the last simulation!
#> int noint
#> costs 53768.32 51510.10
#> lys 9.62 9.62
#> qalys 6.17 6.00
#> ICER NA Inf
#> ICUR NA 13253.26
summary_results_psa(results$output_psa)
#> int noint
#> costs 53768(53768, 53768) 51510(51510, 51510)
#> lys 9.62(9.62, 9.62) 9.62(9.62, 9.62)
#> qalys 6.17(6.17, 6.17) 6(6, 6)
#> ICER NaN(NA, NA) Inf(Inf, Inf)
#> ICUR NaN(NA, NA) 13253(13253, 13253)
psa_ipd <- bind_rows(map(results$output_psa, "merged_df"))
psa_ipd[1:10,] %>%
kable() %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| evtname | evttime | cost | qaly | ly | pat_id | trt | total_costs | total_qalys | total_lys | simulation |
|---|---|---|---|---|---|---|---|---|---|---|
| sick | 0.0000000 | 0.000 | 0.0000000 | 0.0000000 | 1 | int | 46744.30 | 4.180740 | 7.379309 | 1 |
| sicker | 1.6848510 | 6547.811 | 1.3095622 | 1.6369528 | 1 | int | 46744.30 | 4.180740 | 7.379309 | 1 |
| death | 8.5124476 | 40196.491 | 2.8711779 | 5.7423558 | 1 | int | 46744.30 | 4.180740 | 7.379309 | 1 |
| sick | 0.0000000 | 0.000 | 0.0000000 | 0.0000000 | 2 | int | 69943.88 | 5.179030 | 10.144515 | 1 |
| sicker | 0.3581057 | 1423.636 | 0.2847271 | 0.3559089 | 2 | int | 69943.88 | 5.179030 | 10.144515 | 1 |
| death | 12.4769194 | 68520.245 | 4.8943032 | 9.7886065 | 2 | int | 69943.88 | 5.179030 | 10.144515 | 1 |
| sick | 0.0000000 | 0.000 | 0.0000000 | 0.0000000 | 3 | int | 52210.01 | 5.069211 | 8.575177 | 1 |
| sicker | 2.7296539 | 10421.642 | 2.0843283 | 2.6054104 | 3 | int | 52210.01 | 5.069211 | 8.575177 | 1 |
| death | 10.1610637 | 41788.365 | 2.9848832 | 5.9697664 | 3 | int | 52210.01 | 5.069211 | 8.575177 | 1 |
| sick | 0.0000000 | 0.000 | 0.0000000 | 0.0000000 | 4 | int | 49599.19 | 4.793662 | 8.127984 | 1 |
We can also check what has been the absolute number of events per strategy.
| trt | evtname | n |
|---|---|---|
| int | death | 1000 |
| int | sick | 1000 |
| int | sicker | 824 |
| noint | death | 1000 |
| noint | sick | 1000 |
| noint | sicker | 889 |
Plots
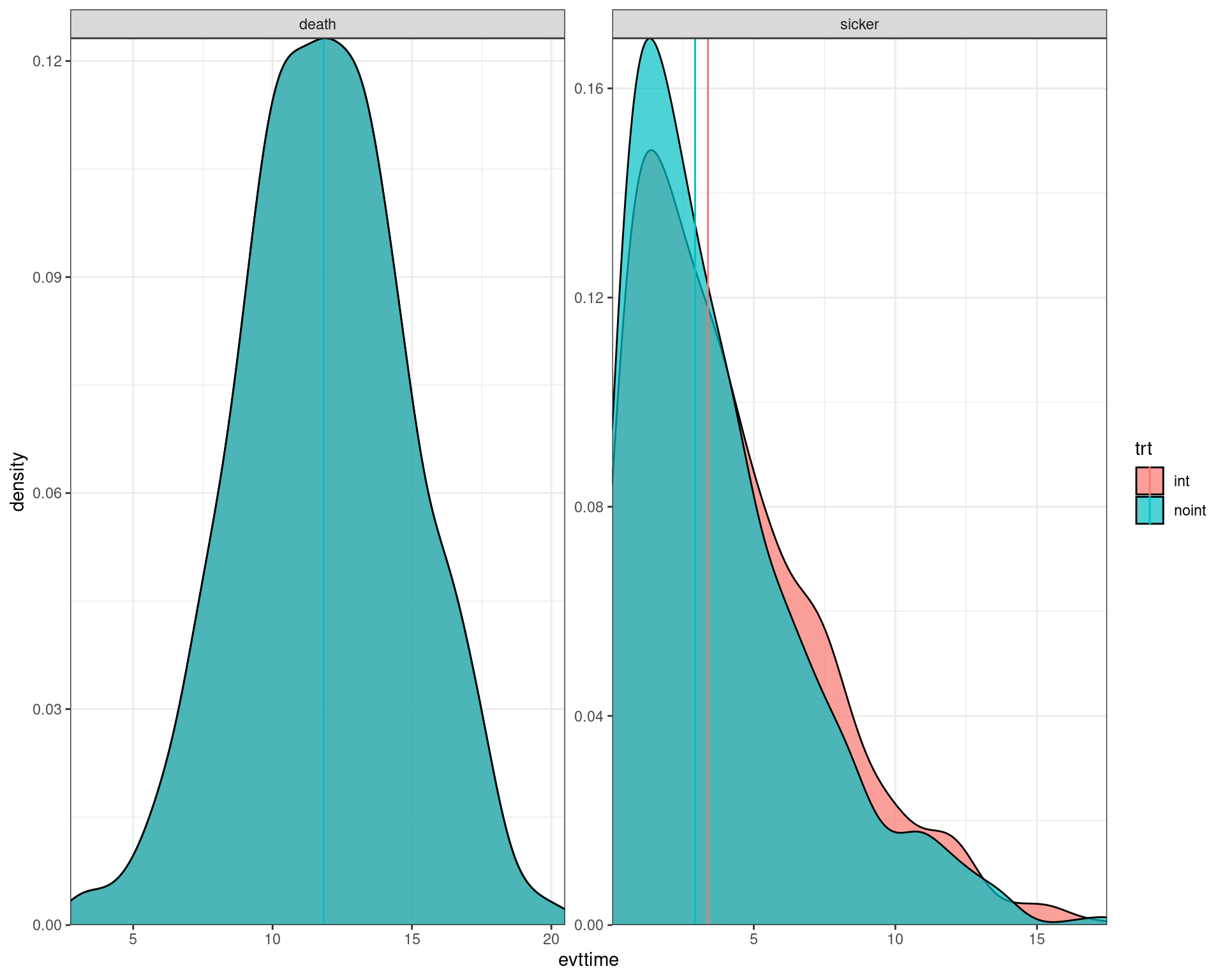
We now use the data output to plot the histograms/densities of the simulation.
data_plot <- results$final_output$merged_df %>%
filter(evtname != "sick") %>%
group_by(trt,evtname,simulation) %>%
mutate(median = median(evttime)) %>%
ungroup()
ggplot(data_plot) +
geom_density(aes(fill = trt, x = evttime),
alpha = 0.7) +
geom_vline(aes(xintercept=median,col=trt)) +
facet_wrap( ~ evtname, scales = "free") +
scale_y_continuous(expand = c(0, 0)) +
scale_x_continuous(expand = c(0, 0)) +
theme_bw()
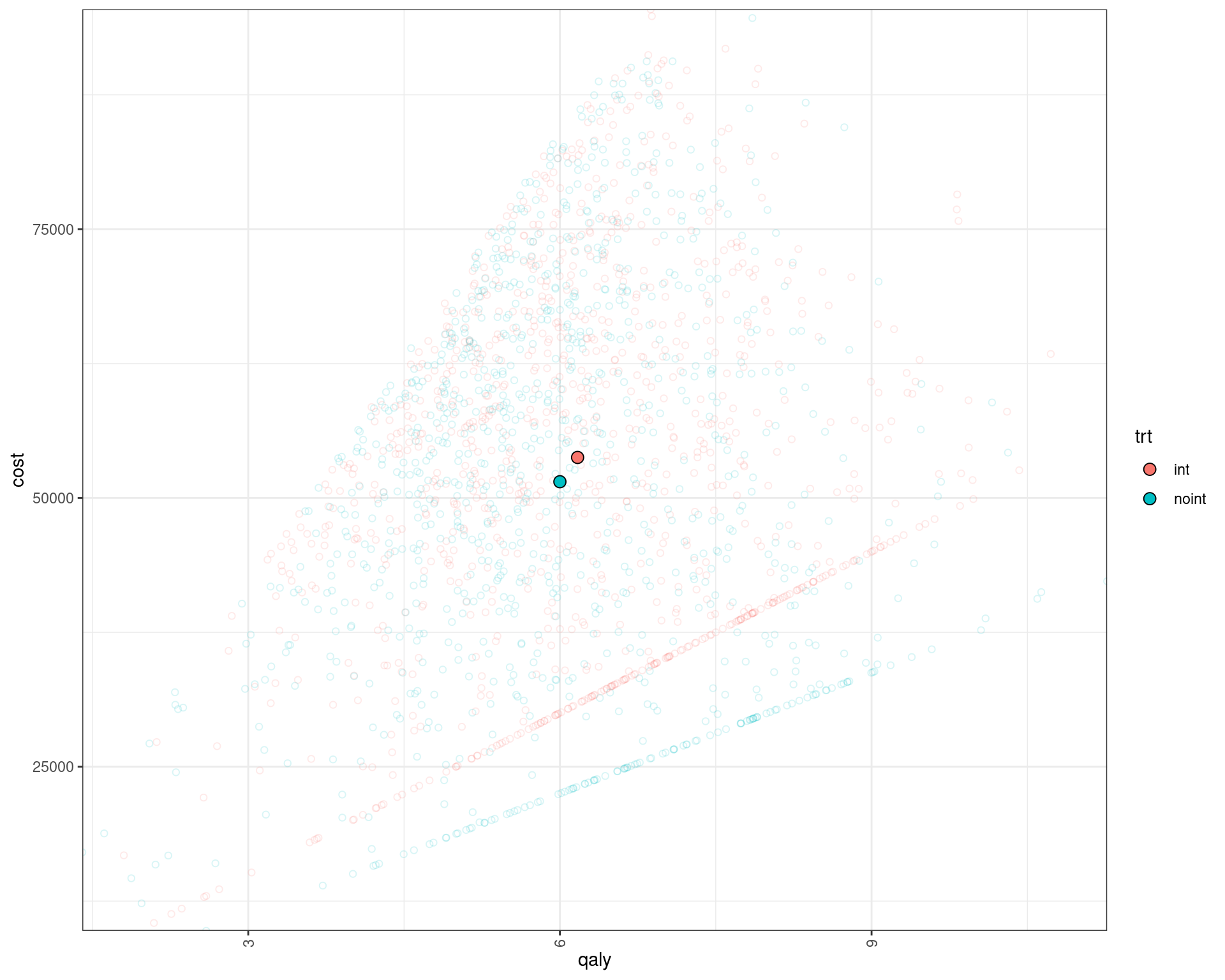
We can also plot the patient level incremental QALY/costs. Note that there are several clusters in the distribution of patients according to their QALY/costs based on the pathway they took (early metastatic vs. remission and cure or recurrence).
data_qaly_cost<- psa_ipd[,.SD[1],by=.(pat_id,trt,simulation)][,.(trt,qaly=total_qalys,cost=total_costs,pat_id,simulation)]
data_qaly_cost[,ps_id:=paste(pat_id,simulation,sep="_")]
mean_data_qaly_cost <- data_qaly_cost %>% group_by(trt) %>% summarise(across(where(is.numeric),mean))
ggplot(data_qaly_cost,aes(x=qaly, y = cost, col = trt)) +
geom_point(alpha=0.15,shape = 21) +
geom_point(data=mean_data_qaly_cost, aes(x=qaly, y = cost, fill = trt), shape = 21,col="black",size=3) +
scale_y_continuous(expand = c(0, 0)) +
scale_x_continuous(expand = c(0, 0)) +
theme_bw()+
theme(axis.text.x = element_text(angle = 90, vjust = .5))